Transformation from/to IR#
In this section, we explain how to transform numerical data to IR.
Poles#
We consider a Green’s function genereated by poles:
where \(\nu\) is a fermionic or bosonic Matsubara frequency. The corresponding specral function \(A(\omega)\) is given by
The modified (regularized) spectral function reads
for the logistic kernel. We immediately obtain
The following code demostrates this transformation for bosons.
import sparse_ir
import numpy as np
%matplotlib inline
import matplotlib.pyplot as plt
beta = 15
wmax = 10
basis_b = sparse_ir.FiniteTempBasis("B", beta, wmax, eps=1e-10)
coeff = np.array([1])
omega_p = np.array([0.1])
rhol_pole = np.einsum('lp,p->l', basis_b.v(omega_p), coeff/np.tanh(0.5*beta*omega_p))
gl_pole = - basis_b.s * rhol_pole
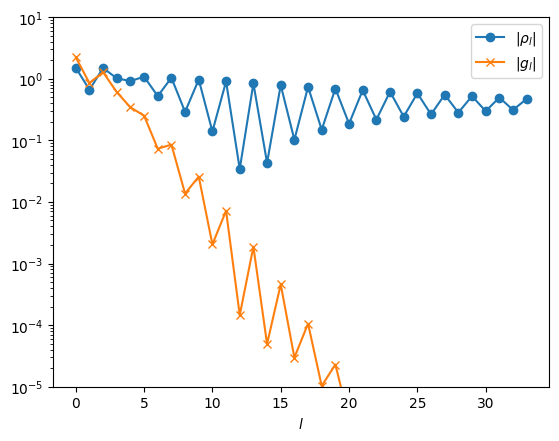
plt.semilogy(np.abs(rhol_pole), marker="o", label=r"$|\rho_l|$")
plt.semilogy(np.abs(gl_pole), marker="x", label=r"$|g_l|$")
plt.xlabel(r"$l$")
plt.ylim([1e-5, 1e+1])
plt.legend()
plt.show()
/opt/hostedtoolcache/Python/3.8.18/x64/lib/python3.8/site-packages/sparse_ir/basis.py:58: UserWarning:
Requested accuracy is 1e-10, which is below the
accuracy 1e-08 for the work data type <class 'numpy.float64'>.
Expect singular values and basis functions for large l to
have lower precision than the cutoff.
Install the xprec package to gain more precision.
sve_result = sve.compute(self._kernel, eps)
Alternatively, we can use spr (sparse pole presentation) module.
from sparse_ir.spr import SparsePoleRepresentation
sp = SparsePoleRepresentation(basis_b, omega_p)
gl_pole2 = sp.to_IR(coeff)
plt.semilogy(np.abs(gl_pole2), marker="x", label=r"$|g_l|$ from SPR")
plt.semilogy(np.abs(gl_pole), marker="x", label=r"$|g_l|$")
plt.xlabel(r"$l$")
plt.ylim([1e-5, 1e+1])
plt.legend()
plt.show()
---------------------------------------------------------------------------
ModuleNotFoundError Traceback (most recent call last)
Cell In[2], line 1
----> 1 from sparse_ir.spr import SparsePoleRepresentation
2 sp = SparsePoleRepresentation(basis_b, omega_p)
3 gl_pole2 = sp.to_IR(coeff)
ModuleNotFoundError: No module named 'sparse_ir.spr'
From smooth spectral function#
For a smooth spectral function \(\rho(\omega)\), the expansion coefficients can be evaluated by computing the integral
One might consider to use the Gauss-Legendre quadrature. As seen in previous sections, the distribution of \(V_l(\omega)\) is much denser than Legendre polynomial \(P_l(x(\tau))\) around \(\tau=0, \beta\). Thus, evaluating the integral precisely requires the use of composite Gauss–Legendre quadrature, where the whole inteval \([-\wmax, \wmax]\) is divided to subintervals and the normal Gauss-Legendre quadrature is applied to each interval. The roots of \(V_l(\omega)\) for the highest \(l\) used in the expansion is a reasonable choice of the division points. If \(\rho(\omega)\) is smooth enough within each subinterval, the result converges exponentially with increasing the degree of the Gauss-Legendre quadrature.
Below, we demonstrate how to compute \(\rho_l\) for a spectral function consisting of of three Gausssian peaks using the composite Gauss-Legendre quadrature. Then, \(\rho_l\) can be transformed to \(g_l\) by multiplying it with \(- S_l\).
# Three Gaussian peaks (normalized to 1)
gaussian = lambda x, mu, sigma:\
np.exp(-((x-mu)/sigma)**2)/(np.sqrt(np.pi)*sigma)
rho = lambda omega: 0.2*gaussian(omega, 0.0, 0.15) + \
0.4*gaussian(omega, 1.0, 0.8) + 0.4*gaussian(omega, -1.0, 0.8)
omegas = np.linspace(-5, 5, 1000)
plt.xlabel(r"$\omega$")
plt.ylabel(r"$\rho(\omega)$")
plt.plot(omegas, rho(omegas))
plt.show()
beta = 10
wmax = 10
basis = sparse_ir.FiniteTempBasis("F", beta, wmax, eps=1e-10)
rhol = basis.v.overlap(rho)
gl = - basis.s * rhol
plt.semilogy(np.abs(rhol), marker="o", label=r"$|\rho_l|$")
plt.semilogy(np.abs(gl), marker="x", label=r"$|g_l|$")
plt.xlabel(r"$l$")
plt.ylim([1e-5, 1])
plt.legend()
plt.show()
\(\rho_l\) is evaluated on arbitrary real frequencies as follows.
rho_omgea_reconst = basis.v(omegas).T @ rhol
plt.xlabel(r"$\omega$")
plt.ylabel(r"$\rho(\omega)$")
plt.plot(omegas, rho(omegas))
plt.show()
From IR to imaginary time#
We are now ready to evaluate \(g_l\) on arbitrary \(\tau\) points. A naive way is as follows.
taus = np.linspace(0, beta, 1000)
gtau1 = basis.u(taus).T @ gl
plt.plot(taus, gtau1)
plt.xlabel(r"$\tau$")
plt.ylabel(r"$G(\tau)$")
plt.show()
Alternatively, we can use TauSampling as follows.
smpl = sparse_ir.TauSampling(basis, taus)
gtau2 = smpl.evaluate(gl)
plt.plot(taus, gtau1)
plt.xlabel(r"$\tau$")
plt.ylabel(r"$G(\tau)$")
plt.show()
From full imaginary-time data#
A numerically stable way to expand \(G(\tau)\) in IR is evaluating the integral
You can use overlap function as well.
def eval_gtau(taus):
uval = basis.u(taus) #(nl, ntau)
return uval.T @ gl
gl_reconst = basis.u.overlap(eval_gtau)
plt.semilogy(np.abs(gl_reconst), label="reconstructed", marker="o")
plt.semilogy(np.abs(gl), label="exact", marker="x")
plt.semilogy(np.abs(gl_reconst - gl), label="error", marker="")
plt.xlabel(r"$l$")
plt.xlabel(r"$|g_l|$")
plt.ylim([1e-20, 1])
plt.legend()
plt.show()