Sparse sampling#
In this page, we describe how to infer IR expansion coefficients using the sparse-sampling techiniques.
import sparse_ir
import numpy as np
%matplotlib inline
import matplotlib.pyplot as plt
plt.rcParams['font.size'] = 15
Setup#
We consider a semicircular spectral modeli (full bandwidth of 2):
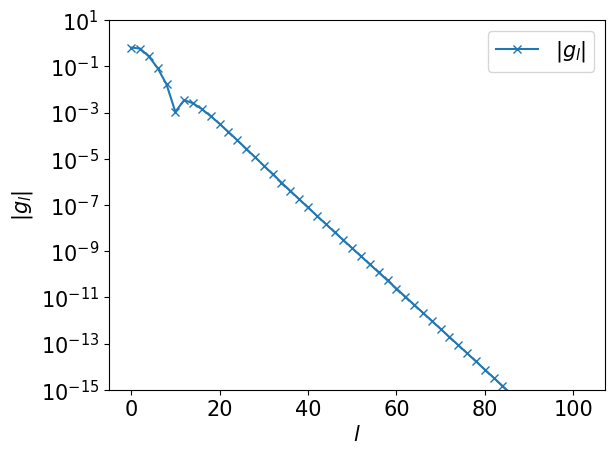
First, we compute the numerically exact expansion coefficients \(g_l\). Below, we plot the data for even \(l\).
def rho(omega):
if np.abs(omega) < 1:
return (2/np.pi) * np.sqrt(1-omega**2)
else:
return 0.0
beta = 10000
wmax = 1
basis = sparse_ir.FiniteTempBasis("F", beta, wmax, eps=1e-15)
rhol = basis.v.overlap(rho)
gl = - basis.s * rhol
ls = np.arange(basis.size)
plt.semilogy(ls[::2], np.abs(gl[::2]), marker="x", label=r"$|g_l|$")
plt.xlabel(r"$l$")
plt.ylabel(r"$|g_l|$")
plt.ylim([1e-15, 10])
plt.legend()
plt.show()
From sampling times#
We first create a TauSampling object for the default sampling times.
smpl_tau = sparse_ir.TauSampling(basis)
print("sampling times: ", smpl_tau.sampling_points)
print("Condition number: ", smpl_tau.cond)
sampling times: [6.47259411e-02 3.41743000e-01 8.43021447e-01 1.57374357e+00
2.54146656e+00 3.75645685e+00 5.23203838e+00 6.98503912e+00
9.03635582e+00 1.14116536e+01 1.41422159e+01 1.72659575e+01
2.08286083e+01 2.48850703e+01 2.95009414e+01 3.47541980e+01
4.07370279e+01 4.75578203e+01 5.53433349e+01 6.42411056e+01
7.44221521e+01 8.60840959e+01 9.94547772e+01 1.14796470e+02
1.32410787e+02 1.52644359e+02 1.75895379e+02 2.02621103e+02
2.33346379e+02 2.68673315e+02 3.09292122e+02 3.55993176e+02
4.09680252e+02 4.71384769e+02 5.42280708e+02 6.23699556e+02
7.17144194e+02 8.24299996e+02 9.47040430e+02 1.08742317e+03
1.24767097e+03 1.43012945e+03 1.63719162e+03 1.87117715e+03
2.13415420e+03 2.42769528e+03 2.75256828e+03 3.10838360e+03
3.49324565e+03 3.90348920e+03 4.33360019e+03 4.77640949e+03
5.22359051e+03 5.66639981e+03 6.09651080e+03 6.50675435e+03
6.89161640e+03 7.24743172e+03 7.57230472e+03 7.86584580e+03
8.12882285e+03 8.36280838e+03 8.56987055e+03 8.75232903e+03
8.91257683e+03 9.05295957e+03 9.17570000e+03 9.28285581e+03
9.37630044e+03 9.45771929e+03 9.52861523e+03 9.59031975e+03
9.64400682e+03 9.69070788e+03 9.73132669e+03 9.76665362e+03
9.79737890e+03 9.82410462e+03 9.84735564e+03 9.86758921e+03
9.88520353e+03 9.90054522e+03 9.91391590e+03 9.92557785e+03
9.93575889e+03 9.94465667e+03 9.95244218e+03 9.95926297e+03
9.96524580e+03 9.97049906e+03 9.97511493e+03 9.97917139e+03
9.98273404e+03 9.98585778e+03 9.98858835e+03 9.99096364e+03
9.99301496e+03 9.99476796e+03 9.99624354e+03 9.99745853e+03
9.99842626e+03 9.99915698e+03 9.99965826e+03 9.99993527e+03]
Condition number: 51.830544391655934
The condition number is around 50, indicating that 1–2 significant digits may be lost in a fit from the sampling times. Let us fit from the sampling times!
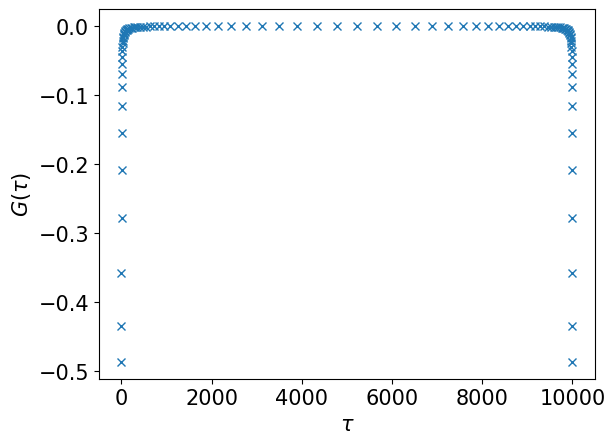
# Evaluate G(τ) on the sampling times
gtau_smpl = smpl_tau.evaluate(gl)
plt.plot(smpl_tau.sampling_points, gtau_smpl, marker="x", ls="")
plt.xlabel(r"$\tau$")
plt.ylabel(r"$G(\tau)$")
plt.show()
# Fit G(τ) on the sampling times
gl_reconst_from_tau = smpl_tau.fit(gtau_smpl)
From sampling frequencies#
We create a MatsubaraSampling object for the default sampling frequencies.
smpl_matsu = sparse_ir.MatsubaraSampling(basis)
print("sampling frequencies: ", smpl_matsu.sampling_points)
print("Condition number: ", smpl_matsu.cond)
sampling frequencies: [-44377 -14981 -8743 -6213 -4765 -3733 -3089 -2523 -2093 -1797
-1513 -1315 -1117 -975 -859 -729 -623 -547 -469 -411
-361 -309 -267 -233 -205 -177 -153 -133 -115 -101
-87 -75 -65 -57 -49 -43 -37 -33 -29 -25
-23 -21 -19 -17 -15 -13 -11 -9 -7 -5
-3 -1 1 3 5 7 9 11 13 15
17 19 21 23 25 29 33 37 43 49
57 65 75 87 101 115 133 153 177 205
233 267 309 361 411 469 547 623 729 859
975 1117 1315 1513 1797 2093 2523 3089 3733 4765
6213 8743 14981 44377]
Condition number: 210.8802635675076
The condition number is slightly larger than that for the sampling times.
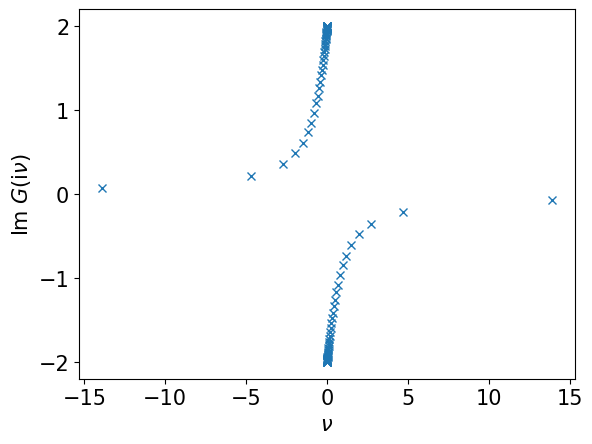
# Evaluate G(iv) on the sampling frequencies
giv_smpl = smpl_matsu.evaluate(gl)
plt.plot((np.pi/beta)*smpl_matsu.wn, giv_smpl.imag, marker="x", ls="")
plt.xlabel(r"$\nu$")
plt.ylabel(r"Im $G(\mathrm{i}\nu)$")
plt.show()
# Fit G(τ) on the sampling times
gl_reconst_from_matsu = smpl_matsu.fit(giv_smpl)
Comparison with exact results#
We now compare the reconstructed expansion coefficients with the exact one. For clarity, we plot only the data for even \(l\).
plt.semilogy(ls[::2], np.abs(gl[::2]), marker="", ls="-", label="Exact")
plt.semilogy(ls[::2], np.abs(gl_reconst_from_tau[::2]), marker="x", label="from sampling times")
plt.semilogy(ls[::2], np.abs(gl_reconst_from_matsu[::2]), marker="+", label="from sampling frequencies")
plt.xlabel(r"$l$")
plt.ylabel(r"$|g_l|$")
plt.ylim([1e-17, 10])
plt.legend(frameon=False)
plt.show()
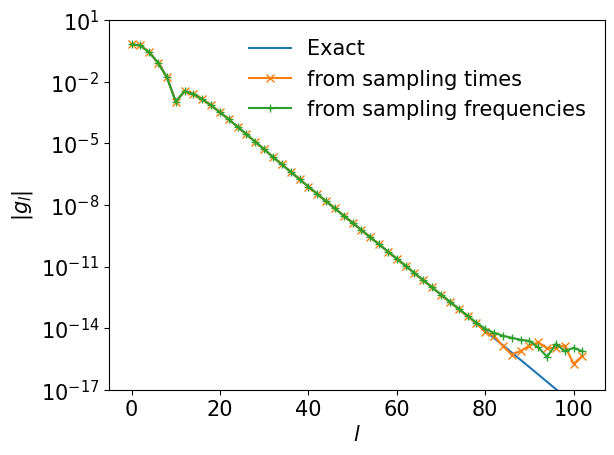
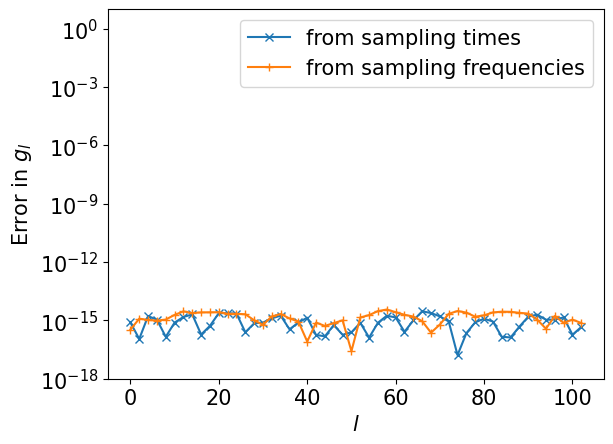
We saw a perfect match! Let us plot the differences from the exact one.
plt.semilogy(ls[::2], np.abs((gl_reconst_from_tau-gl)[::2]), marker="x", label="from sampling times")
plt.semilogy(ls[::2], np.abs((gl_reconst_from_matsu-gl)[::2]), marker="+", label="from sampling frequencies")
plt.xlabel(r"$l$")
plt.ylabel(r"Error in $g_l$")
plt.ylim([1e-18, 10])
plt.legend()
plt.show()